dTOX Documentation
1 Login:(same as dREG)
The user needs to log in by clicking 'Log in' link at the top-right corner of the page. Having an account provides a number of benefits, and is free and easy.
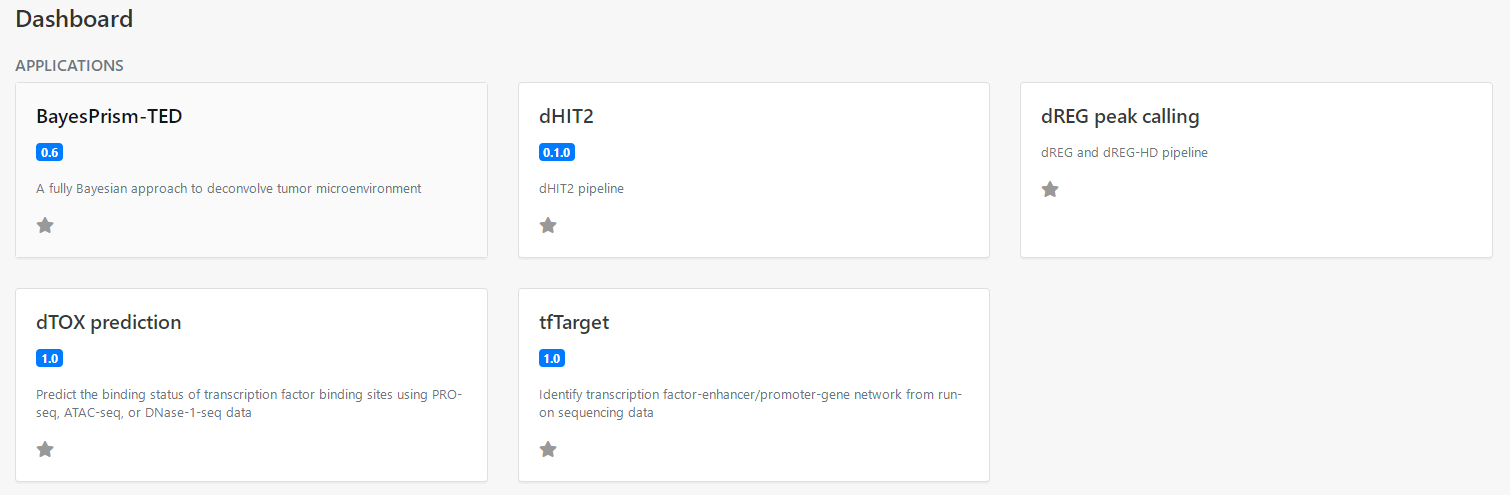
2 Create a new experiment
Select the dTOX application on the Dashboard panel to create a data analysis for your data, as the following screenshot (Figure 2).
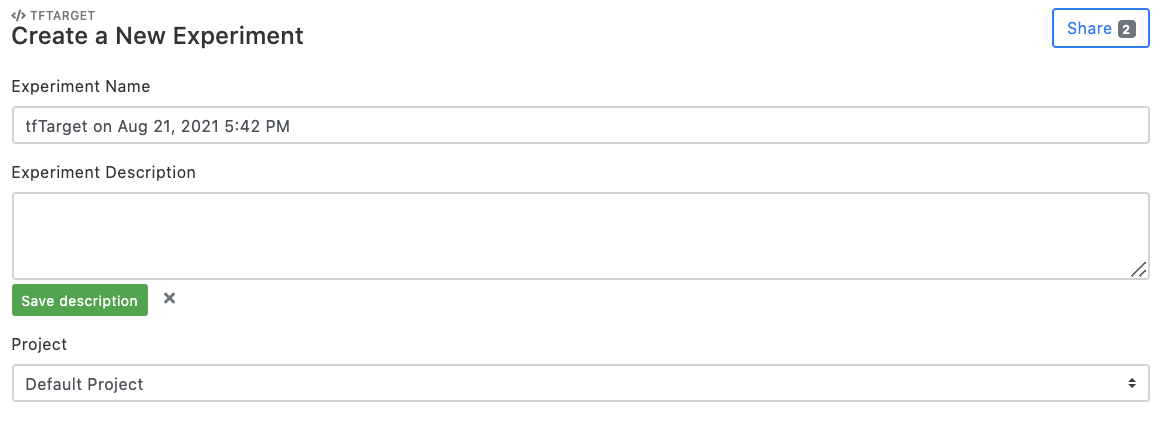
3 Set experiment name
Set "Experiment Name", and click "Add a description" to comment on the experimental setup page (optional). Choose the project that the experiment belongs to. By default, the "Default Project" is created and used.
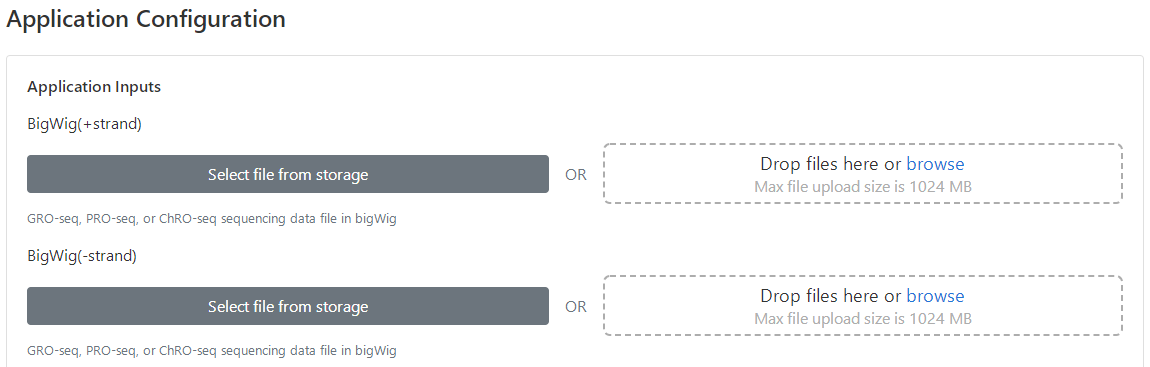
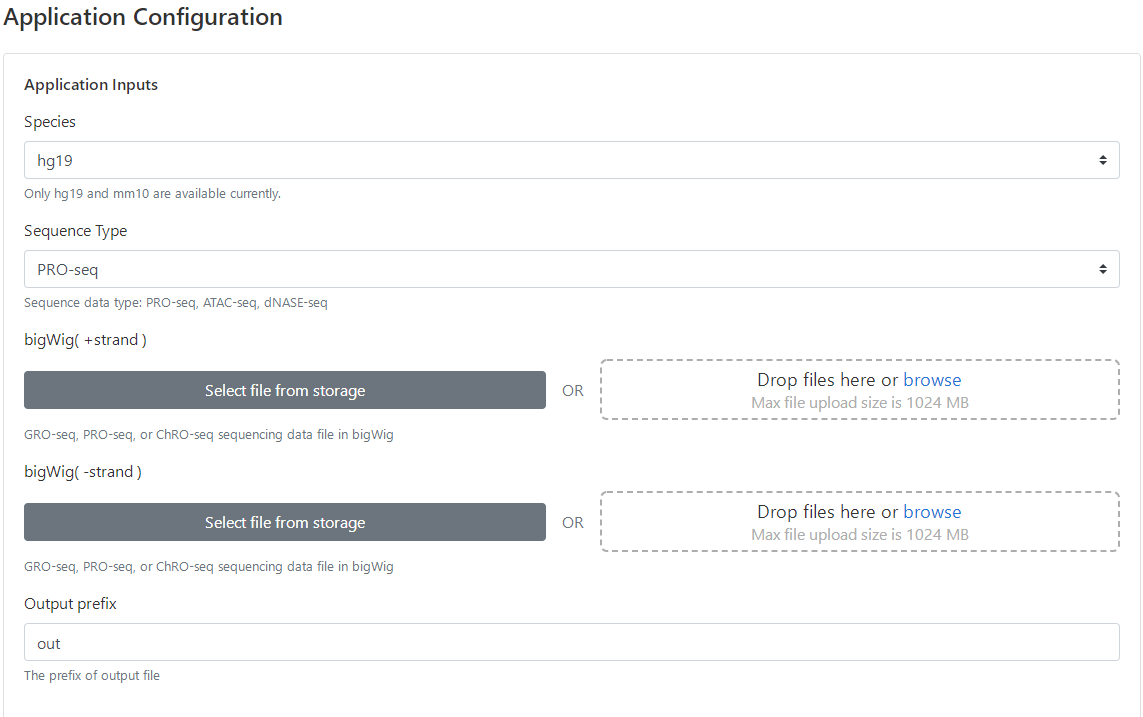
4 Upload bigWig files
There are two ways users may use to upload bigWigs.
(1) Click "Select files from storage" to choose existing files submitted for previous tasks, or
(2) click "Drop files here or browse" to upload new files from user's storage. Note that the bigWig files of run-on sequencing are strand-specific, and hence the ordering of bigWig files needs to be matched for plus and minus strands within each condition. Additionally, as dREG uses DESeq2 to model differential transcription and perform hypothesis testing, at least two replicates are required for each condition.
5 Set computing parameters
(1) Specify the genome assembly of bigWig files. Information of genome assemblies is required for defining gene bodies for quantifying their transcriptional activity and computing motif enrichment scores. Currently, only hg19 and mm10 are supported. If additional genome assemblies are needed, please submit a request to the admin.
(2) Specify the type of RO-seq experiment.
(3) Specify the prefix of the output files. This can help distinguish results from multiple experiments.
6 Submit the job
Once steps 1-5 are finished, proceed to "Save and launch". Input data and parameters will be submitted to the computing node of the XSEDE cluster via the dTOX gateway server. Click the checkbox next to "Receive email notification of experiment status" if needed. dTOX peak calling will be automatically performed on bigWigs merged for each condition. Upon launching, users will be directed to the "Experiments" page, shown in Fig. 4. A typical experiment usually finishes within 4 hrs. Users may view the progress by logging in and clicking the "Experiment" button on the left control panel at the dashboard.
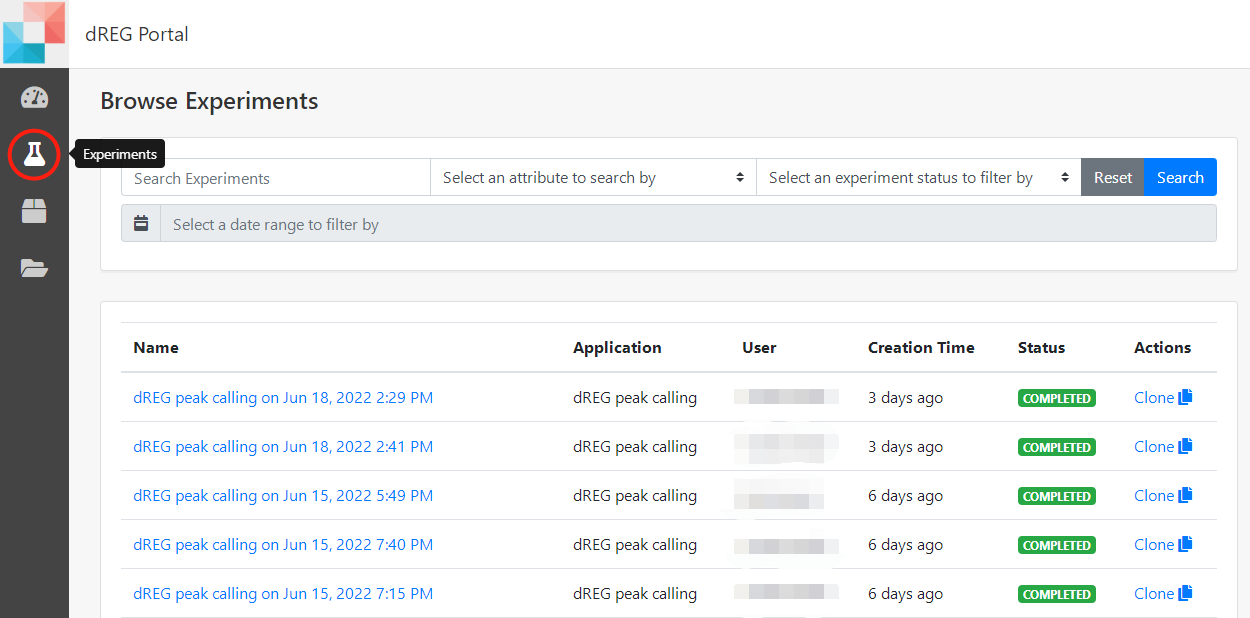
7 Check the status
Users may view the progress by logging in and clicking the "Experiments" button on the left control panel at the dashboard. All experiments submitted are listed on this page.
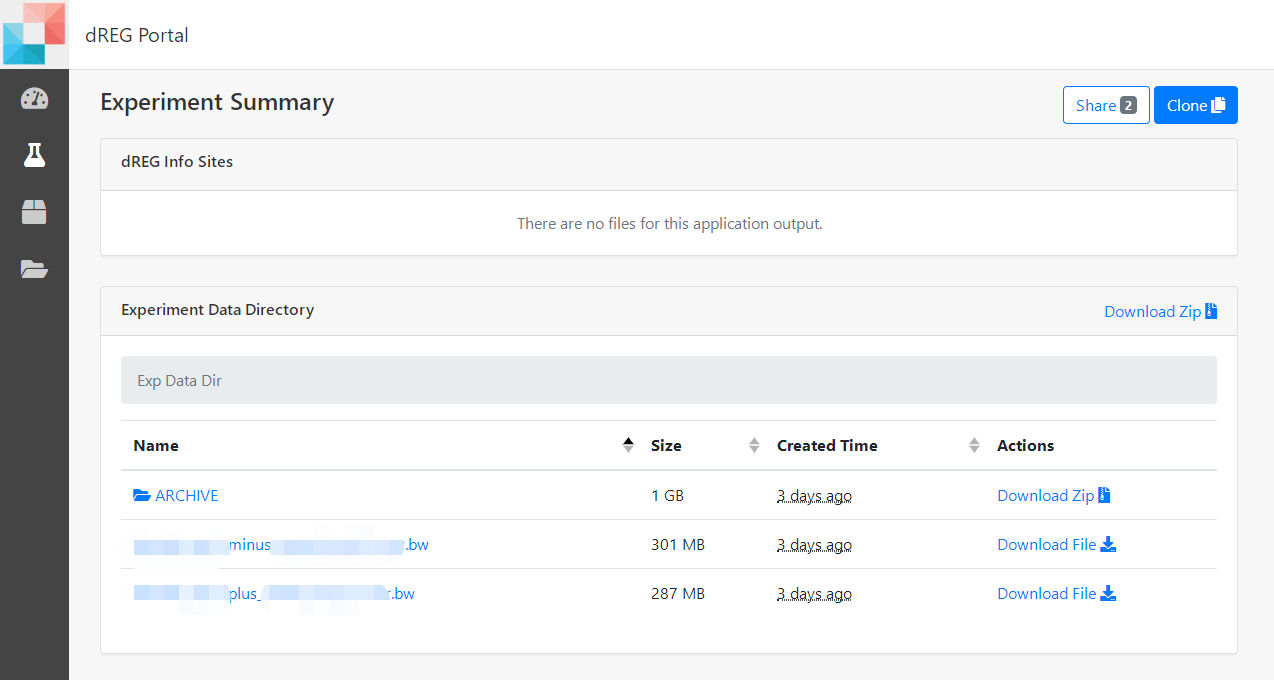
8 Check the results
Once a job is completed, the user can click selected dREG experiment and the website will jump to Experiment Summary page. All parameters used to set up the experiment are listed on this page. The user can also access output files of dREG stored in the ARCHIVE. Just click the ARCHIVE to check any single result file. A compressed file, including input bigWigs file set, two task log files and all result files, is also provided for users. Click Download Zip button to download a compressed file. The downloaded file with the 'tar.gz' extension can be decompressed by the 'tar' command, the file with the 'gz' extension can be decompressed by the 'gunzip' command in Linux.
In Safari, it could be problematic because Safari tries to unzip the compressed results automatically using a non-compatible compress method. Please check this link to disable this feature.
The input to dTOX consists of two bigWig files which represent either the position of RNA polymerase on the positive and negative strands (PRO-seq) or the accessibility on the positive and negative strands (DNase-I-seq or ATAC-seq). The sequence alignment and processing steps to make the input bigWig files are a major factor influencing how accurately dTOX predicts transcription factor binding.
A key component of all datatypes is that data represents unnormalized raw counts. dTOX assumes that data represents the number of individual sequence tags that are located at each genomic position. For this reason, it is critical that input data is not normalized. The server checks to ensure that input data is expressed as integers, and will return an error if this is not the case.
Users can also use scripts generated in the Danko lab to create compatible bigWig files. Options for scripts at different starting points in the analysis are given below:
- Convert raw fastq files into bigWig.
Our pipeline produces bigWig files that are compatible with dTOX, and can be found at the following URLs: https://github.com/Danko-Lab/proseq_2.0 (PRO-seq), https://github.com/Danko-Lab/atac (ATAC-seq), https://github.com/Danko-Lab/dnase (DNase-I-seq). The pipelines automate routine pre-processing and alignment steps, including pre-processing reads to remove the adapter sequences and trim based on base quality, and deduplicate the reads if UMI barcodes are used. Sequencing reads are mapped to a reference genome using BWA. Aligned BAM files are converted into bigWig format in which each read is represented by a single base.
- Convert mapped reads in BAM files into bigWigs.
We provide scripts that convert mapped reads from a BAM file into bigWig files that are compatible with dTOX. The scripts are avavailable on our GitHub page. For PRO-seq: https://github.com/Danko-Lab/RunOnBamToBigWig. For DNase-I-seq: https://github.com/Danko-Lab/utils/dnase/BamToBigWig. For ATAC-seq: https://github.com/Danko-Lab/utils/atacseq/BamToBigWig.
Other considerations:
The quality and quantity of the experimental data are major factors in determining how sensitive dTOX will be in detecting transcription factor binding. To increase the number of reads available for transcription factor binding detection, we encourage users to merge biological replicates in order to improve statistical power prior to running dTOX. Additionally, to compare binding predictions between conditions we recommend comparing samples at similar sequencing depths or down sampling to create similar sequencing depths.
We have found that visualizing aligned data in a genome browser prior (e.g., IGV or UCSC) to downstream analysis is a useful way to catch any data quality or alignment issues.
1) A dTOX run generates a compressed file including the following files:
| File name | Description |
|---|---|
| $PREFIX.dTOX.bound.bed.gz | TFBS regions that are predicted as bound. The file includes chromosome, start, ending, MOTIF ID, RTFBSDB score, strand, dTOX score, bound status. Decompress it with 'gunzip' in Linux. |
Informative position: Loci denoted as "informative positions" meet the following criteria: contain more than 1 reads in 400 bp interval on either strand. Informative positions are used to predict transcription factor binding.
dTOX decision value: Training and prediction is done using a Support Vector Regression model where a label of 1 indicates transcription factor binding. The predicted values from the pre-trained model are called dTOX decision values. A dTOX decision value close to 1 indicates that a position likely to be bound.
The dTOX output file contains the binding status of our entire set of motifs with PWMs. To find the binding status of the motifs you are interested in, you can run our R script that extracts the Motif IDs that belong to a particular transcription factor. The script is located here. This script requires 3 arguments: the name of the file with the dTOX results, the transcription factor you want to extract, and an output file name. To run this script on Unix or Linux, you need to use the following command:
R --vanilla --slave --args out.dTOX.bound.bed.gz TF outputFile.bed.gz < extract-bound-TF.R
2) In the Web storage folder there are some files required by the WashU genome browser:
| File name | Description |
|---|---|
| $PREFIX.dTOX.bound.bw | The bigWig file converted from bound motifs ($PREFIX.dTOX.bound.bed.gz). |
| *.bed.gz.tbi | The index files generated from the corresponding bed files. Please ignore them if you download the results. |
3) There are one log file in the Web storage folder:
| File name | Description |
|---|---|
| slurm-??????.out | The verbose log output of dTOX package. |
dREG Gateway is online service that supports Web-based science through the execution of online computational experiments and the management of data. Below are frequent questions about the dREG Gateway and the dTOX program.
Q: How should I prepare bigWig files for use with dTOX?
A: Information about how to prepare files can be found on the Danko lab github page here for PRO-seq , DNase , and ATAC-seq .
Q: How should I do when I meet the computational failure in the dREG gateway?
A: There are two types of error you may have, we explain how to identify your error and how to handle it here.
Q: Which browser works well with the dREG gateway?
A: We have tested in the Firefox, Google Chrome and Safari so far. For IE (version 10 or 11) and some version of Safari, you maybe have trouble showing sequence data in WashU genome browser. For Safari users, please read next Q&A.
Q: What should the Safari users be aware of?
A: By default, Safari unzips a zip file automatically when you download it. However dTOX results are compressed by the 'bgzip' command which is not compatiable with the Safari method. It would be problematic when you download dTOX results. Please refer to this link to disable this feature in Safari and then download the compressed results from dREG gateway. Secondly, when you click the genome browser link, please use the Left-Click, don't use Right-Click menu and the menu option "open a new tab".
Q: Will dTOX work with my data type?
A: dTOX was trained and tested on PRO-seq, ATAC-seq, and DNase-I-seq. dTOX will also work well with data collected by any run-on and sequencing method, including GRO-seq, PRO-seq, or ChRO-seq. Other methods that map the location of RNA polymerase genome wide using alternative tools (for example, NET-seq) will most likely work well, but are not officially supported.
Q: Will the pre-trained models work using data from my species?
A: Models are currently available only in mammalian organisms. The length and density of genes, which vary considerably between highly divergent species, affects the way that a transcribed promoter or enhancer looks. For this reason, models can only be used in species. We are working to create models in widely-used model organisms, including drosophila and C. elegans.
Q: How deeply do I need to sequence PRO-seq libraries?
A: Sensitivity is reasonable at ~40 million mapped reads and saturates at ~100 million mapped reads. See our analysis here: supplementary figure 3 in dREG paper.
Q: How long do my data and results keep in the dREG gateway?
A: One month.
Q: How do I cite dTOX?
A: Please cite our papers if you use dTOX results in your publication:
(1) ADD CITATION. Choate, L. A., Wang, Z., & Danko, C. G. (2018). Identification of transcription factor binding using genome-wide accessibility and transcription. bioRxiv.
Q: Do I have to create account before using this service?
A: Yes, this system is supported by an NSF funded supercomputing resource known as XSEDE, who regularly needs to report bulk usage statistics to NSF. Nevertheless, data that you provide are completely safe.
Q: How do I know the status of the computational nodes?
A: Since we can't update this web site very often, the gateway status is updated here on the dREG page based on the notifications of the XSEDE community.
Q: Who do I thank for the computing power?
A: This web-based tool is powered by SciGaP and Apache Airavata and the GPU servers are supported by the XSEDE.
Q: I have another question that is not on this FAQ. How can I contact you?
A: Yes, please contact us with any questions! Zhong(zw355 at cornell.edu). Charles(cgd24 at cornell.edu).



