The dREG gateway is a cloud platform that hosts two bioinformatics online services for functional analysis of sequencing data, dREG transcriptional regulatory element peak calling and dTOX transcription factor binding prediction. These online services are developed by the Danko lab at the Baker Institute and Cornell University and supported by the SciGap (Science Gateway Platform as a Service) and XSEDE (Extreme Science and Engineering Discovery Environment).
The gateway is built on the infrastructure services of SciGaP and the computional resources of XSEDE as the following architecture, the details can be found in this paper: "Building a Science Gateway For Processing and Modeling Sequencing Data Via Apache Airavata". In PEARC 2018: Practice and Experience in Advanced Research Computing, July 22-26, 2018, Pittsburgh, PA, USA.
SciGaP supports access to core infrastructure services required by Science Gateways, including: user identity, accounts, authorization, and access to multiple computational resources from XSEDE.
XSEDE provides GPU resources under the project (TG-BIO160048 and TG-BIO180027: dREG Science Gateway).
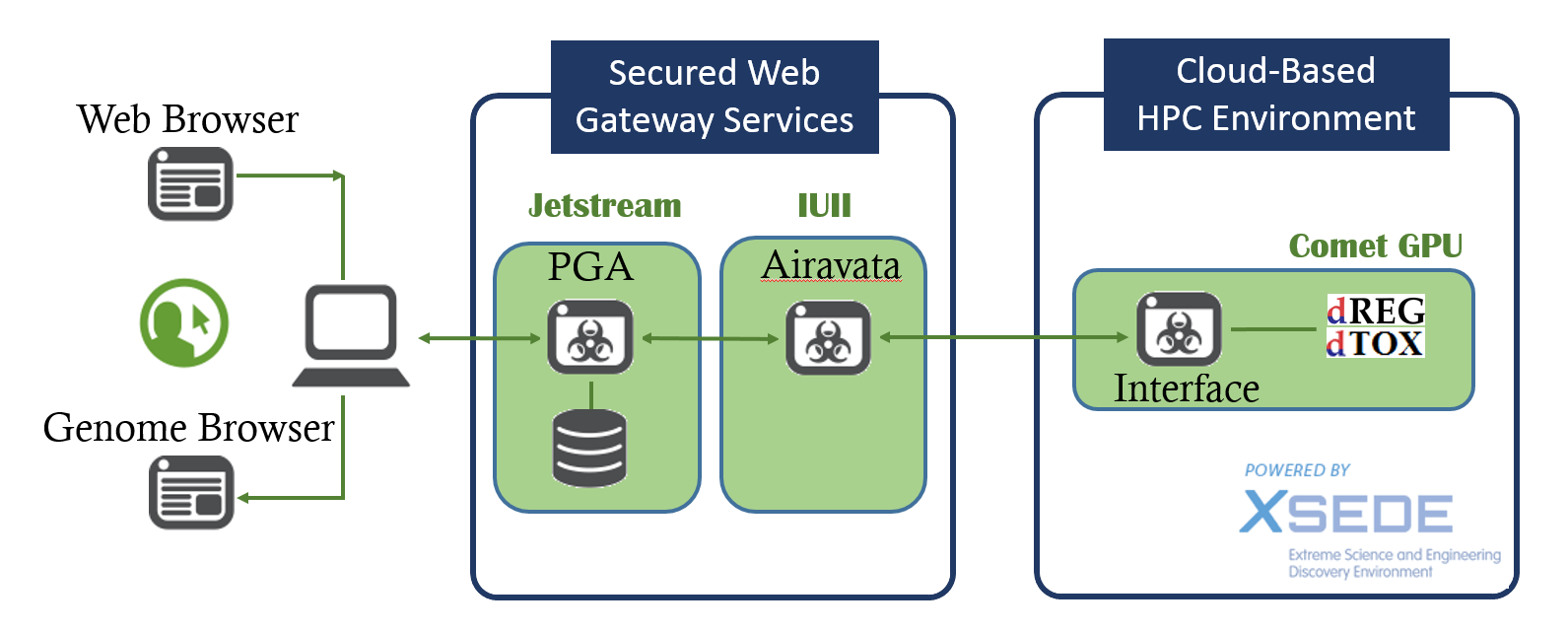
Gateway Architecture
Danko Lab at Cornell University studies how gene expression programs are encoded in mammalian DNA sequences, and how these 'regulatory codes' contribute to evolution, development, and disease. Our specialty is developing statistics and machine-learning approaches to analyze genomic sequencing data, prepared using DNase-I-seq, ATAC-seq, PRO-seq, RNA-seq, and related assays. Our tools borrow a wide variety of ideas from the fields of statistics and machine-learning, including recent uses of hidden Markov models, support vector machines, and artificial neural networks
Apache Airavata is a software framework that enables you to compose, manage, execute, and monitor large scale applications and workflows on distributed computing resources such as local clusters, supercomputers, computational grids, and computing clouds.
PGA(PHP Gateway with Airavata) is a framework to build science gateway with the Airavata API. dREG gateway is built on top of PGA by modifying it.